 |
StarVine
|
 |
StarVine
|
Multi-variate data class. More...
Public Member Functions | |
| def | __init__ (self, mvdData=pd.DataFrame(), mvdWeights=pd.DataFrame()) |
| def | setData (self, dataDict, weights=None) |
| Collect data from dictionary with {str: np_1darray} {key, value} pairs into a pandas dataFrame. | |
| def | plot (self, kwargs) |
| generate pairwise scatter plots | |
| def | setUVD (self, uvdList) |
| Collect uni-variate data sets into a multivariate data object. More... | |
| def | computeKDEpdf (self, bandwidth=None) |
| Computes mulitvariate kernel density function. More... | |
| def | computeCov (self, weighted=False) |
| computes cov matrix More... | |
| def | computePC (self) |
| Computes principal components of multivariate data set. More... | |
| def | computePCOP (self, retainFracVar=0.95, reducedDim=None) |
| Computes a projection matrix. More... | |
| def | applyPCA (self) |
| Provides a reduced order view of the current MVD object. More... | |
| def | plotExplainedVar (self) |
| Plots fractional explained varience as a function of number of principal components retained. | |
Public Attributes | |
| uvdPool | |
| mvdData | |
| mvdDataWeights | |
| nDims | |
| mvdKDEpdf | |
| mvdCov | |
| eig_pairs | |
| frac_explained_var | |
| cum_frac_explained_var | |
| pcW | |
Multi-variate data class.
Performs principal component analysis to reduce the dimensionality of a large data set.
| def starvine.mvar.mvd.Mvd.applyPCA | ( | self | ) |
Provides a reduced order view of the current MVD object.
| def starvine.mvar.mvd.Mvd.computeCov | ( | self, | |
weighted = False |
|||
| ) |
computes cov matrix
| weighted | bool if True, utilizes mvdDataWeights (vols or areas) as frequency weights, essentially counting samples which represent more "area" or "volume" in the domain more times. True by default. |
| def starvine.mvar.mvd.Mvd.computeKDEpdf | ( | self, | |
bandwidth = None |
|||
| ) |
Computes mulitvariate kernel density function.
A kernel density function is constructed by combining many locally supported "mini PDFs". This is a data smoothing operation.
| <b>float</b> | (optional) bandwidth. Default is to use the "scott" factor: 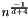 |
| def starvine.mvar.mvd.Mvd.computePC | ( | self | ) |
Computes principal components of multivariate data set.
Provides a measure of explained varience per principal component, the principal compenent directions and magnitudes.
| def starvine.mvar.mvd.Mvd.computePCOP | ( | self, | |
retainFracVar = 0.95, |
|||
reducedDim = None |
|||
| ) |
Computes a projection matrix.
Maps from the original data space to a reduced param space.
| retainFracVar | double Desired fraction of explained varience to retain |
| reducedDim | int Target reduced data dimension |
| def starvine.mvar.mvd.Mvd.setUVD | ( | self, | |
| uvdList | |||
| ) |
Collect uni-variate data sets into a multivariate data object.
| uvdList | list of uvar.Uvd instances |