 |
StarVine
|
 |
StarVine
|
Univariate parameterized model base class. More...
Public Member Functions | |
| def | __init__ (self, paramsString, momtype, bounds, args, kwargs) |
| def | setupGMM (self, data, w=1.0, nMoM=None) |
| General Method of Moments setup. More... | |
| def | fitKDE (self, data, args) |
| Fit gaussian kernel density fn to data. | |
| def | setLogPrior (self, logPriorFn) |
| Set log of prior distribution
. More... | |
| def | setupMCMC (self, nwalkers, params, data, wgts=None, args) |
| Initilize the MCMC sampler. More... | |
| def | fitMCMC (self, iters, pos=None) |
| Execute MCMC model parameter fitting. More... | |
| def | fitMLE (self, data, params0, weights=None, args, kwargs) |
| Generic maximum (log)likelyhood estimate of paramers. More... | |
Public Attributes | |
| nParams | |
| nMomentConditions | |
| internalGMM | |
| kde | |
| logPriorFn | |
| walker_pos | |
| nwalkers | |
| sampler | |
Private Member Functions | |
| def | _pdf (self, x, args) |
| Pure virtual pdf model function. More... | |
| def | _logPosterior (self, params, data, wgts) |
Posterior distribution 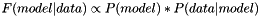
. | |
| def | _paramCheck (input_f) |
| Parameter bounds checker. More... | |
| def | _loglike (self, params, data, wgts) |
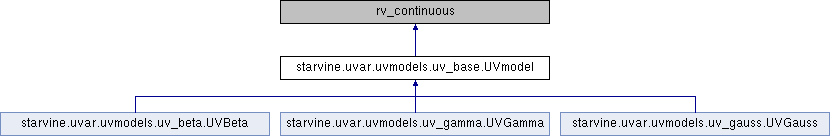
Univariate parameterized model base class.
Extends the capabilities of the rv_continuous base class of the scipy.stats module with the GMM capabilites of statmodels.sandbox.regression.gmm (general method of moments fitting).
With the combined capability of scipy and statmodels, it is easy to obtain maximum likelyhood and MoM estimators for univariate model parameters.
Methods inherited from scipy.stats.rv_continuous:
rvs(*args, **kwds) Random variates of given type.
pdf(x, *args, **kwds) Probability density function at x of the given RV.
logpdf(x, *args, **kwds) Log of the probability density function at x of the given RV.
cdf(x, *args, **kwds) Cumulative distribution function of the given RV.
logcdf(x, *args, **kwds) Log of the cumulative distribution function at x of the given RV.
sf(x, *args, **kwds) Survival function (1 - cdf) at x of the given RV.
logsf(x, *args, **kwds) Log of the survival function of the given RV.
ppf(q, *args, **kwds) Percent point function (inverse of cdf) at q of the given RV.
isf(q, *args, **kwds) Inverse survival function (inverse of sf) at q of the given RV.
moment(n, *args, **kwds) n-th order non-central moment of distribution.
stats(*args, **kwds) Some statistics of the given RV.
entropy(*args, **kwds) Differential entropy of the RV.
expect([func, args, loc, scale, lb, ub, ...]) Calculate expected value of a
function with respect to the distribution.
median(*args, **kwds) Median of the distribution.
mean(*args, **kwds) Mean of the distribution.
std(*args, **kwds) Standard deviation of the distribution.
var(*args, **kwds) Variance of the distribution.
interval(alpha, *args, **kwds) Confidence interval with equal areas around the median.
__call__(*args, **kwds) Freeze the distribution for the given arguments.
fit(data, *args, **kwds) Return MLEs for shape, location, and scale parameters from data.
fit_loc_scale(data, *args) Estimate loc and scale parameters from data using 1st and 2nd moments.
nnlf(theta, x) Return negative loglikelihood function.
|
private |
| params | array-like model parameters |
| data | np_array input data |
| wgts | np_array input data weights |
|
private |
Parameter bounds checker.
|
private |
Pure virtual pdf model function.
Note: the number of paramers is "infered" from the signature of the _pdf OR _cdf methods.
| def starvine.uvar.uvmodels.uv_base.UVmodel.fitMCMC | ( | self, | |
| iters, | |||
pos = None |
|||
| ) |
Execute MCMC model parameter fitting.
Must call setupMCMC before calling this method.
| iters | int number of MCMC iterations |
| pos | Initial position of walkers |
| def starvine.uvar.uvmodels.uv_base.UVmodel.fitMLE | ( | self, | |
| data, | |||
| params0, | |||
weights = None, |
|||
| args, | |||
| kwargs | |||
| ) |
Generic maximum (log)likelyhood estimate of paramers.
| data | Input data to fit to |
| params0 | Initial guess for model parameters |
| weights | Input data weights |
Note: compare to scipy's model.fit() method results. Scipy's model.fit() method does not allow for weights :(
| def starvine.uvar.uvmodels.uv_base.UVmodel.setLogPrior | ( | self, | |
| logPriorFn | |||
| ) |
Set log of prior distribution
![\[ ln(P(\theta)) \]](form_24.png)
.
| logPriorFn | function logPrior |
| def starvine.uvar.uvmodels.uv_base.UVmodel.setupGMM | ( | self, | |
| data, | |||
w = 1.0, |
|||
nMoM = None |
|||
| ) |
General Method of Moments setup.
| data | Input univariate data |
| w | Data weights |
Example General Method of Moments use:
>>> my_uvmodel = UVmodel(...) >>> params0 = np.array([2.0, 1.0]) >>> my_uvmodel.internalGMM.fit(params0, maxiter=2, optim_method='nm', wargs=dict(centered=False))
| def starvine.uvar.uvmodels.uv_base.UVmodel.setupMCMC | ( | self, | |
| nwalkers, | |||
| params, | |||
| data, | |||
wgts = None, |
|||
| args | |||
| ) |
Initilize the MCMC sampler.
| nwalkers | int Number of walkers |
| params | list of doubles Model parameter list |
| data | np_array data array |
| wgts | np_array data weights |